This document is relevant for: Inf1, Inf2, Trn1, Trn2, Trn3
Capture and Launch a Profile in Neuron Explorer#
This guide covers how to capture a profile, launch the Neuron Explorer, use the Profile Manager, and view Neuron Explorer in your IDE.
Note
This guide currently only covers Neuron device profiling. For users interested in system profiling, refer to Neuron Profiler 2.0 (Beta) User Guide. The new Neuron Explorer UI will support an integrated system profiling experience in a future release.
Capturing profiles#
To get a better understanding of our workload’s performance, you must collect the raw device traces and runtime metadata in the form of an NTFF (Neuron Trace File Format) which you can then correlate with the compiled NEFF (Neuron Executable File Format) to derive insights.
Set the following environment variables before compiling to capture more descriptive layer names and stack frame information.
export XLA_IR_DEBUG=1
export XLA_HLO_DEBUG=1
For NKI developers, set NEURON_FRAMEWORK_DEBUG in addition to the two above to enable kernel source code tracking:
export NEURON_FRAMEWORK_DEBUG=1
If profiling was successful, you will see NEFF (.neff) and NTFF (.ntff) artifacts in the specified output directory similar to the following:
output
└── i-0ade06f040a13f2bf_pid_210229
├── 395760075800974_instid_0_vnc_0.ntff
└── neff_395760075800974.neff
Device profiles for the first execution of each NEFF per NeuronCore are captured, and NEFF/NTFF pairs with the same prefix (for PyTorch) or unique hash (for JAX or CLI) must be uploaded together. See the section on uploading profiles for more details.
Capturing a profile with PyTorch#
The context-managed profiling API in torch_neuronx.experimental.profiler allows you to profile specific blocks of code. To use the profiling API, import it into your application:
from torch_neuronx.experimental import profiler
Then, profile a block of code using the following code:
with torch_neuronx.experimental.profiler.profile(
profile_type='operator',
target='neuron_profile',
output_dir='./output') as profiler:
Full code example:
import os
import torch
import torch.nn as nn
import torch.nn.functional as F
# XLA imports
import torch_xla
import torch_xla.core.xla_model as xm
import torch_xla.debug.profiler as xp
import torch_neuronx
from torch_neuronx.experimental import profiler
# Global constants
EPOCHS = 2
# Declare 3-layer MLP Model
class MLP(nn.Module):
def __init__(self, input_size = 10, output_size = 2, layers = [5, 5]):
super(MLP, self).__init__()
self.fc1 = nn.Linear(input_size, layers[0])
self.fc2 = nn.Linear(layers[0], layers[1])
self.fc3 = nn.Linear(layers[1], output_size)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return F.log_softmax(x, dim=1)
def main():
# Fix the random number generator seeds for reproducibility
torch.manual_seed(0)
# XLA: Specify XLA device (defaults to a NeuronCore on Trn1 instance)
device = xm.xla_device()
# Start the proflier context-manager
with torch_neuronx.experimental.profiler.profile(
profile_type='operator',
target='neuron_profile',
output_dir='./output') as profiler:
# IMPORTANT: the model has to be transferred to XLA within
# the context manager, otherwise profiling won't work
model = MLP().to(device)
optimizer = torch.optim.SGD(model.parameters(), lr=0.01)
loss_fn = torch.nn.NLLLoss()
# start training loop
print('----------Training ---------------')
model.train()
for epoch in range(EPOCHS):
optimizer.zero_grad()
train_x = torch.randn(1,10).to(device)
train_label = torch.tensor([1]).to(device)
#forward
loss = loss_fn(model(train_x), train_label)
#back
loss.backward()
optimizer.step()
# XLA: collect ops and run them in XLA runtime
xm.mark_step()
print('----------End Training ---------------')
if __name__ == '__main__':
main()
Capturing a profile with JAX#
When using the JAX context-managed profiling API, set two extra environment variables to signal the profile plugin to begin capturing device profile data when the profiling API is invoked.
os.environ["NEURON_RT_INSPECT_DEVICE_PROFILE"] = "1"
os.environ["NEURON_RT_INSPECT_OUTPUT_DIR"] = "./output"
Then, profile a block of code:
with jax.profiler.trace(os.environ["NEURON_RT_INSPECT_OUTPUT_DIR"]):
Full code example:
from functools import partial
import os
import jax
import jax.numpy as jnp
from jax.sharding import Mesh, NamedSharding, PartitionSpec as P
from jax.experimental.shard_map import shard_map
from time import sleep
from functools import partial
os.environ["NEURON_RT_INSPECT_DEVICE_PROFILE"] = "1"
os.environ["NEURON_RT_INSPECT_OUTPUT_DIR"] = "./output"
jax.config.update("jax_default_prng_impl", "rbg")
mesh = Mesh(jax.devices(), ('i',))
def device_put(x, pspec):
return jax.device_put(x, NamedSharding(mesh, pspec))
lhs_spec = P('i', None)
lhs = device_put(jax.random.normal(jax.random.key(0), (128, 128)), lhs_spec)
rhs_spec = P('i', None)
rhs = device_put(jax.random.normal(jax.random.key(1), (128, 16)), rhs_spec)
@jax.jit
@partial(shard_map, mesh=mesh, in_specs=(lhs_spec, rhs_spec),
out_specs=rhs_spec)
def matmul_allgather(lhs_block, rhs_block):
rhs = jax.lax.all_gather(rhs_block, 'i', tiled=True)
return lhs_block @ rhs
with jax.profiler.trace(os.environ["NEURON_RT_INSPECT_OUTPUT_DIR"]):
out = matmul_allgather(lhs, rhs)
for i in range(10):
with jax.profiler.TraceAnnotation("my_label"+str(i)):
out = matmul_allgather(lhs, rhs)
sleep(0.001)
expected = lhs @ rhs
with jax.default_device(jax.devices('cpu')[0]):
equal = jnp.allclose(jax.device_get(out), jax.device_get(expected), atol=1e-3, rtol=1e-3)
print("Tensors are the same") if equal else print("Tensors are different")
Capturing a profile from CLI#
In certain cases, you may want to profile the application without requiring code modifications such as when deploying a containerized application through EKS. Note that when capturing with the CLI, profiling will be enabled for the entire lifetime of the application. If more granular control is required for profiling specific sections of the model, it is recommended to use the PyTorch or JAX APIs.
To enable profiling without code change, run your workload with the following environment variables set:
export NEURON_RT_INSPECT_ENABLE=1
export NEURON_RT_INSPECT_DEVICE_PROFILE=1
export NEURON_RT_INSPECT_OUTPUT_DIR=./output
python train.py
Setting up the Neuron Explorer UI#
Use the neuron-explorer tool from aws-neuronx-tools to start the UI and API servers that are required for viewing profiles.
neuron-explorer view
By default, the UI will be launched on port 3001 and the API server will be launched on port 3002.
If this is launched on a remote EC2 instance, use port-forwarding to enable local viewing of the profiles.
ssh -i <key.pem> <user>@<ip> -L 3001:locahost:3001 -L 3002:localhost:3002
Browser UI#
After the above setup, navigate to localhost:3001 in the browser to view the Profile Manager.
VSCode Extension#
The UI is also available as a VSCode extension, enabling better native integration for features such as code linking.
First, download the Visual Studio Code Extension (.vsix) file from aws-neuron/aws-neuron-sdk.
Get the Neuron Explorer VSCode Extension
Once downloaded, open the command palette by pressing CMD+Shift+P (MacOS) or Ctrl+Shift+P (Windows), type > Extensions: Install from VSIX... and press Enter. When you are prompted to select a file, select neuron-explorer-2.27.0.vsix and then the Install button (or press Enter) to install the extension.
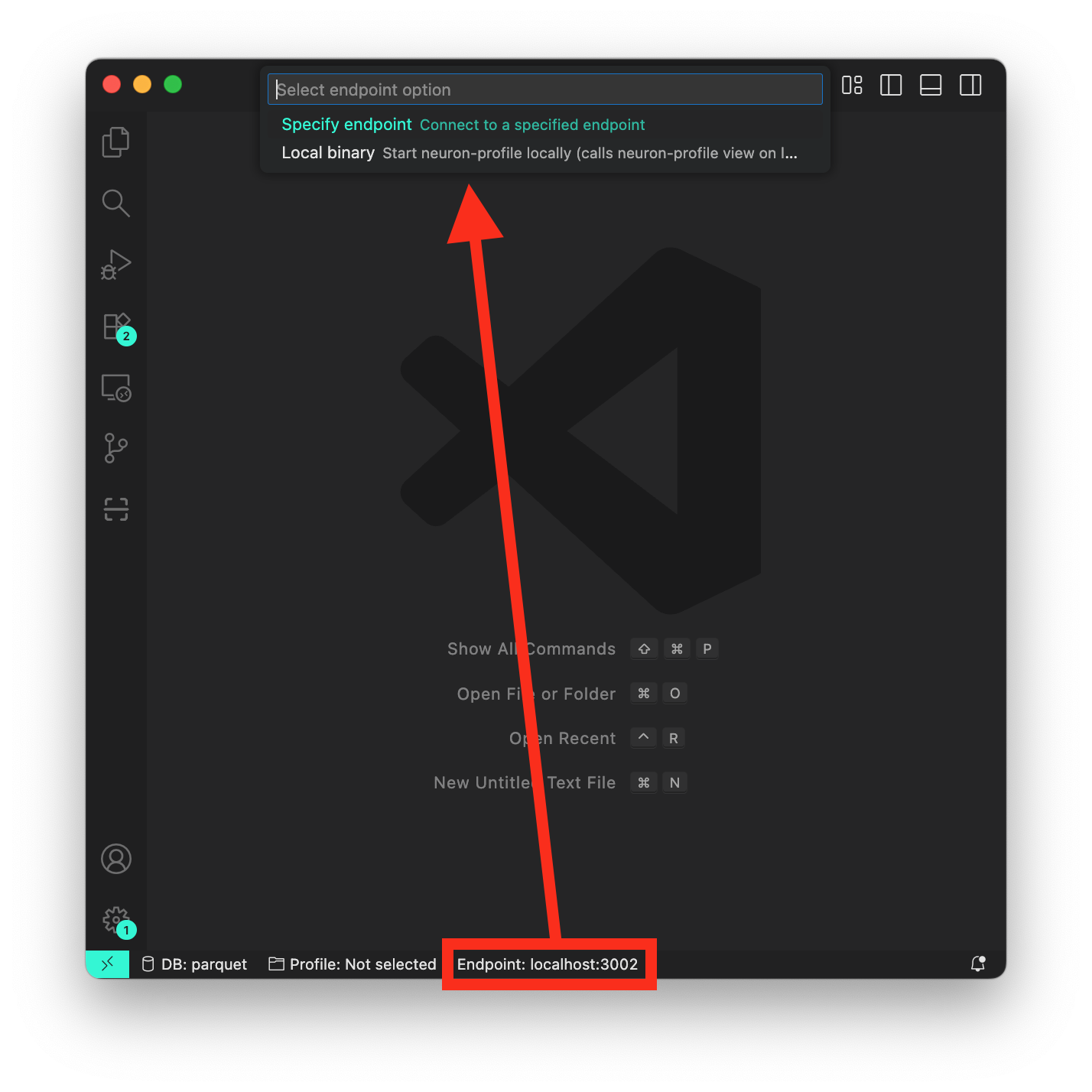
Ensure the SSH tunnel is established by following the steps above. Otherwise, specify a custom endpoint by selecting the extension in the left activity bar. Then, navigate to the “Endpoint” action on the bottom bar of your VSCode session and select “Custom endpoint”, and enter localhost:3002.
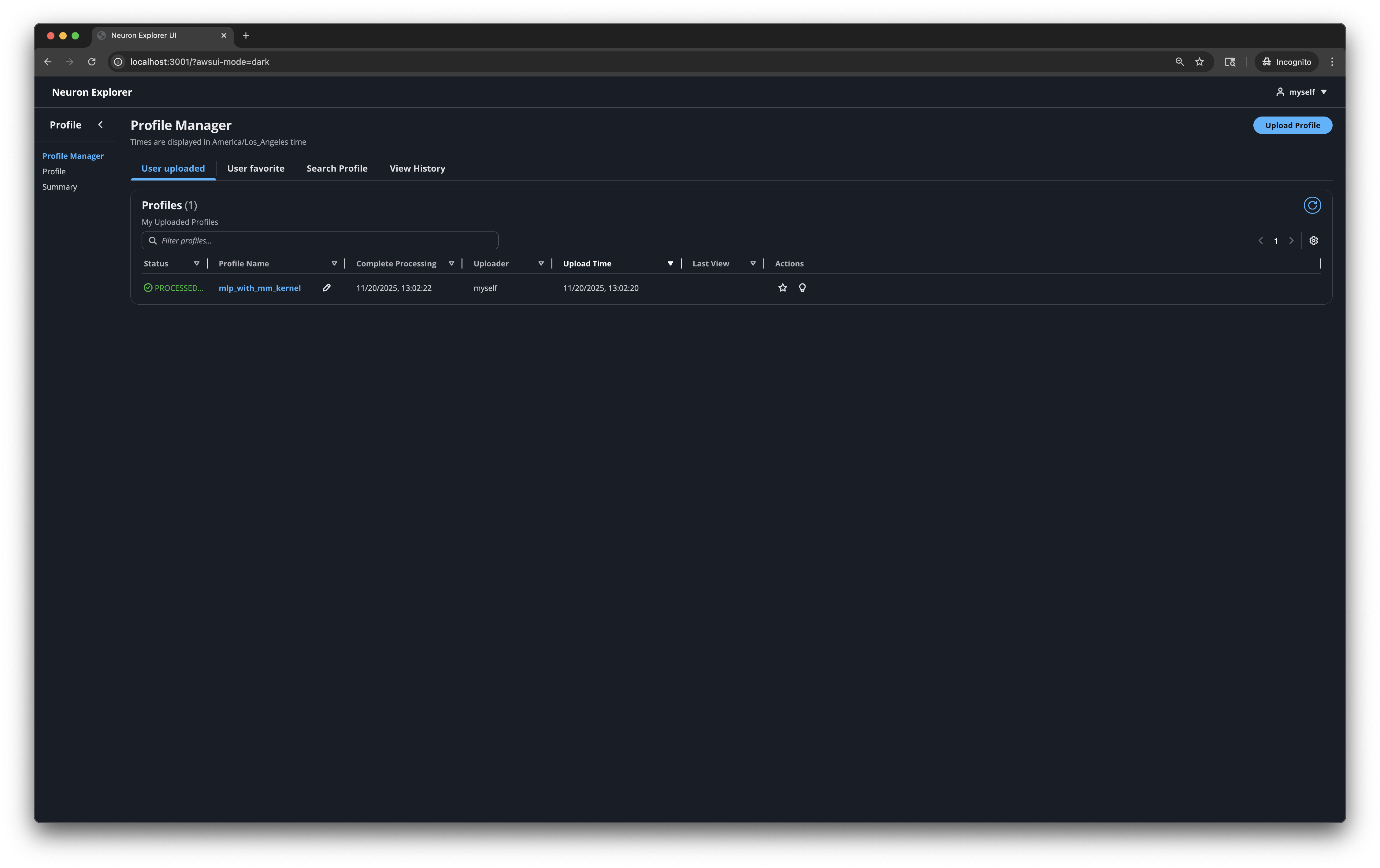
From there, navigate to the Profile Manager page through the extension UI in the left activity bar.
Profile Manager#
Profile Manager is a page for uploading artifact (NEFF, NTFF and source code) and selecting profiles to access.
Uploading a profile#
Click on “Upload Profile” to select NEFF, NTFF, and source code folders from file system.
Note
“Profile name” is a required field. You cannot upload a profile with existing name unless the option “Force Upload” is checked at the bottom. Force Upload currently will overwrite the existing profile with the same name.
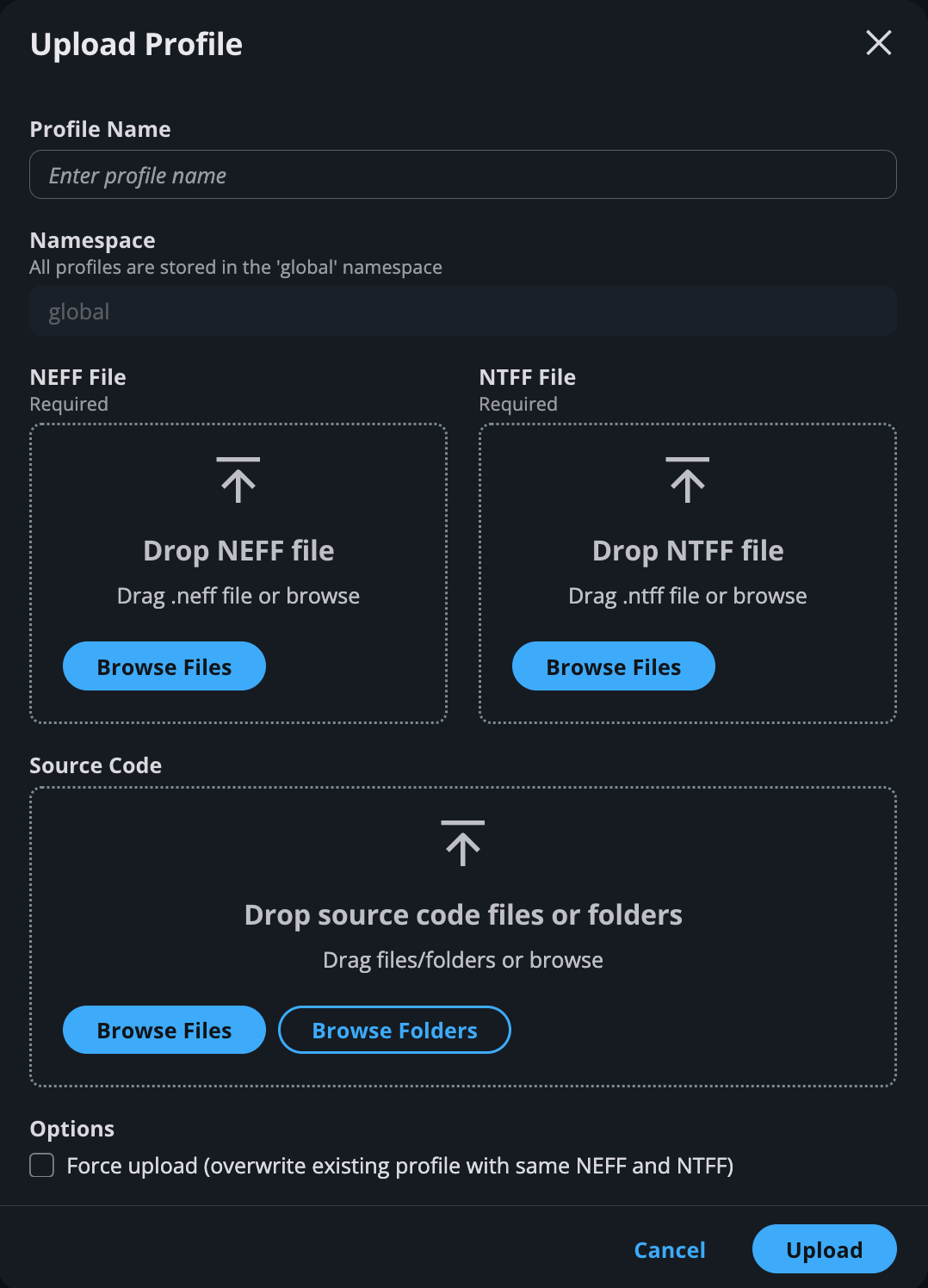
Uploading source code#
After uploading a profile, the processing task is shown under “User Uploaded”. The “Refresh” button on the top-right can be used to check whether the processing is completed.
Note
For uploading source code, the UI only supports the upload of folders, individual files, or compressed files in the gzipped tar .tar.gz archive format.
Listing profiles#
All uploaded profiles are provided in the Profile Manager page with details such as the processing status and upload time, along with various quick access actions.
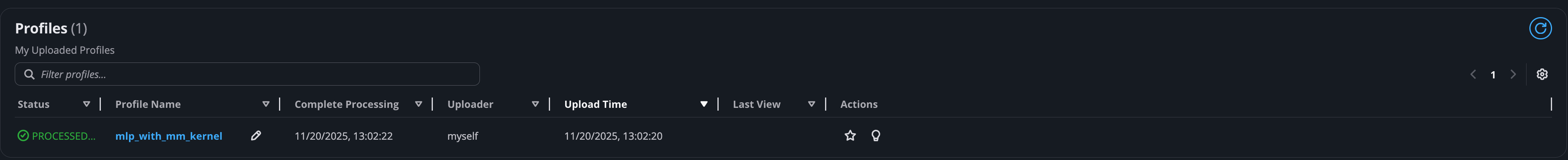
Pencil button: Rename a profile.
Star button: Mark this profile as favorite profile. This profile will be shown in the User’s favorites list.
Bulb button: Navigate to the summary page of this profile. For more details on the summary page, see this overview of the Neuron Explorer Summary Page.
Clicking on the name of profile takes you to its corresponding profile page.
This document is relevant for: Inf1, Inf2, Trn1, Trn2, Trn3
