This document is relevant for: Inf2, Trn1, Trn2, Trn3
Pipeline Parallelism Overview#
Pipeline parallelism is a technique used in deep learning model training to improve efficiency and reduce the training time of large neural networks. Currently NeuronxDistributed’s pipeline parallelism is built specially for transformer based models, where each Neuron core will be assigned with a subset of transformer layers. Pipelining is a technique to achieve true parallelization in pipeline parallelism, by having the Neuron cores compute simultaneously on different data samples, and to overcome the performance loss due to sequential computation. When you use pipeline parallelism, training job is executed in a pipelined fashion over microbatches to maximize device usage.
Model partitioning#
In NeuronxDistributed, we use Pytorch’s FX to trace the model and do partition on the FX IR. User simply needs to specify where to cut the pipeline stages, and our algorithm will cut the pipeline stages and assign the corresponding modules to each Neuron core automatically. Currently we require user to provide model partition decision but auto-partition will be supported in the future. Here is an example of simple partition with 5 linear layers
# original NN module
class MyModule(torch.nn.Module):
def __init__(self):
super().__init__()
self.linears = torch.nn.ModuleList([torch.nn.Linear(4, 4) for _ in range(5)])
def forward(self, x):
for lin in self.linears:
x = lin(x)
return x
m = MyModule()
gm = torch.fx.symbolic_trace(m)
print(gm)
"""
GraphModule(
(linears): Module(
(0): Linear(in_features=4, out_features=4, bias=True)
(1): Linear(in_features=4, out_features=4, bias=True)
(2): Linear(in_features=4, out_features=4, bias=True)
(3): Linear(in_features=4, out_features=4, bias=True)
(4): Linear(in_features=4, out_features=4, bias=True)
)
)
def forward(self, x):
linears_0 = getattr(self.linears, "0")(x); x = None
linears_1 = getattr(self.linears, "1")(linears_0); linears_0 = None
linears_2 = getattr(self.linears, "2")(linears_1); linears_1 = None
linears_3 = getattr(self.linears, "3")(linears_2); linears_2 = None
linears_4 = getattr(self.linears, "4")(linears_3); linears_3 = None
return linears_4
"""
If user decide to cut the pipeline stage at the 3nd linear call, after partition there will be 2 submodules, where submod_0 contains first 3 linear layers and submod_1 contains last 2 linear layers.
After Split module
GraphModule(
(submod_0): GraphModule(
(linears_0): Linear(in_features=4, out_features=4, bias=True)
(linears_1): Linear(in_features=4, out_features=4, bias=True)
(linears_2): Linear(in_features=4, out_features=4, bias=True)
)
(submod_1): GraphModule(
(linears_3): Linear(in_features=4, out_features=4, bias=True)
(linears_4): Linear(in_features=4, out_features=4, bias=True)
)
)
def forward(self, x):
submod_0 = self.submod_0(x); x = None
submod_1 = self.submod_1(submod_0); submod_0 = None
return submod_1
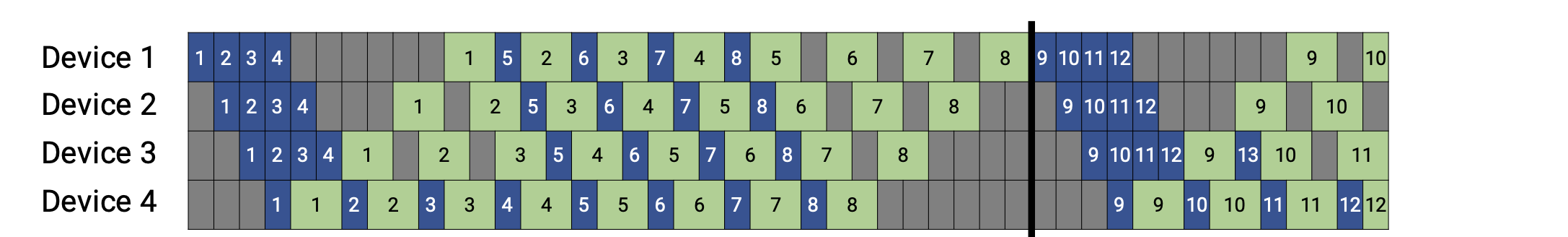
Pipeline Execution Schedule#
Pipelining is based on splitting a mini-batch into microbatches, which are fed into the training pipeline one-by-one and follow an execution schedule defined by the library runtime. A microbatch is a smaller subset of a given training mini-batch. The pipeline schedule determines which microbatch is executed by which device for every time slot.
For example, depending on the pipeline schedule and the model partition, Neuron core i might perform (forward or backward) computation on microbatch b while Neuron core i+1 performs computation on microbatch b+1, thereby keeping both Neuron cores active at the same time. An example taken from Megatron paper is showed as below
This document is relevant for: Inf2, Trn1, Trn2, Trn3
